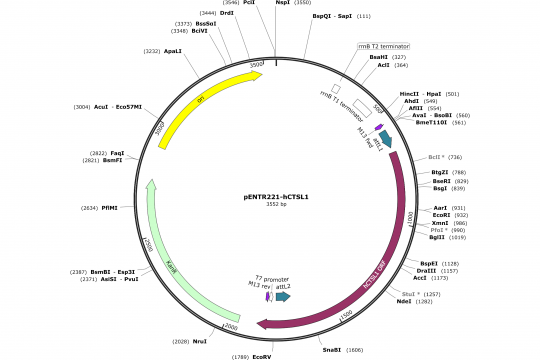
Assembly of an OCAA collection Entry vector (pENTR221)
This article is for people, who do molecular cloning. More specifically for people who need to deal with Gateway vectors. Let's assume you received a Gateway clone from somebody. You know the insert sequence and you know the backbone. One of the most common backbones is pENTR221. Let take as an example insert the human CTSL1 cDNA, more specifically the clone id 100010639 from the OCAA clone collection. You know the insert sequence from its Accession Number (BC012612).
You want the full DNA sequence of this vector in order to be able to use smart cloning software like SnapGene to help you with your cloning design. But this software requires you to have the full sequence of the construct (or at least the full sequence of its important parts). So how do you figure out the full sequence of the pENTR221-CTSL1 clone?
The insert sequence you can get from NCBI: Just type in the Accession number that you got for your clone. Download the sequence in Fasta or Genbank format.
Very interestingly, the otherwise smart SnapGene software does not know the pENTR221 vector. So you need to google the vector backbone "pENTR221 DNA sequence". You get many hits and here are just four of them:
1. http://dnasu.org/DNASU/GetVectorDetail.do?vectorid=279
2. https://plasmid.med.harvard.edu/PLASMID/GetVectorDetail.do?vectorid=279
3. https://www.genomics-online.com/vector-backbone/48/pentr221/
4. http://yrgene.com/documents/vector/pentr221.pdf
Matthias, the author of the 4th source gives the sequence in a PDF file, which is not advisable. If you copy the DNA sequence from this file, it will be all scrambled up, because PDF does not read the groupings of 10 nucleotides line-by-line. Dear Matthias, do not use a PDF for distributing or documenting DNA sequences! If you MUST do so, please attach a plain text file of the nucleotide sequence to the PDF! Of course, you can extract the DNA sequence with a smart PDF tool like PDF Studio Pro in the correct order. For some strange reason, the sequence of the 3rd URL deviates from the other four being the only one that has the full attachment sites (attL1 and attL2). However, it does not matter which one of the sequences you use for the assembly, because the differences are all in the area that is removed during the assembly process (I don't know how the pENTR221 vector was prepared for the library cloning of my specific example, but it looks to me that the original vector was opened with a single DraI digest (which creates blunt ends) and then first the linker were added and thereafter the insert.
I suggest you use the sequence from the 3rd URL (because it is in Fasta format) and import it into SnapGene and let SnapGene detect common features. Now you still need the linker. How do you know which linker have been used? We get most of our Gateway clones from an in-house replica of the OCAA clone collection and you can download the full list of clones as an Excel spreadsheet from here. The data includes for each clones the linker that have been used (there are 8 different 5'-linker and 16 different 3'-linker). Unfortunately, it seems to be that this Excel sheet contains some errors, because some linker contain a stop codon, but are nevertheless marked "without stop" and vice-versa.
For our example clone the following linker have been used:
5'-linker:
GTACAAAAAAGCAGGCTCCACCATG
3'-linker:
TAGGACCCAGCTTTCTTGTAC
Almost all of the 5'-linker contain the Kozak sequence (CACC) as the last nucleotides before the insert starts and a few contain in addition to the Kozak the ATG itself (like the one above). At the other end of the linker you can easily identify the homologous sequence with the end of the attL1 of the pENTR221 backbone (GTACAAAAAAG).
The 3'-linker are more heterogenous but they all contain the CTTTCTTG sequence from the attL2. When they are used to make clones with a stop codon, then they all start with that very stop codon (TAG, TGA or TAA).
Now you just need to copy the open reading frame from your insert sequence in between the linker sequences. If your 3'-linker contains the initiation-ATG, you need to skip it. Also do not copy the stop codon, because in the "with stop codon clones" it is always included in the linker and in the "without stop codon clones" you don't want to have it. For our example this sequence comprises nucleotides 202-1197 of Accession Number BC012612. That would be:
AATCCTACACTCATCCTTGCTGCCTTTTGCCTGGGAATTGCCTCAGCTACTCTAACATTTGATCACAGTTTAGAGGC
ACAGTGGACCAAGTGGAAGGCGATGCACAACAGATTATACGGCATGAATGAAGAAGGATGGAGGAGAGCAGTGTGGGAGA
AGAACGTGAAGATGATTGAACTGCACAATCAGGAATACAGGGAAGGGAAACACAGCTTCACAATGGCCATGAACGCCTTT
GGAGACATGACCAGTGAAGAATTCAGGCAGGTGATGAATGGCTTTCAAAACCGTAAGCCCAGGAAGGGGAAAGTGTTCCA
GGAACCTCTGTTTTATGAGGCCCCCAGATCTGTGGATTGGAGAGAGAAAGGCTACGTGACTCCTGTGAAGAATCAGGGTC
AGTGTGGTTCTTGTTGGGCTTTTAGTGCTACTGGTGCTCTTGAAGGACAGATGTTCCGGAAAACTGGGAGGCTTATCTCA
CTGAGTGAGCAGAATCTGGTAGACTGCTCTGGGCCTCAAGGCAATGAAGGCTGCAATGGTGGCCTAATGGATTATGCTTT
CCAGTATGTTCAGGATAATGGAGGCCTGGACTCTGAGGAATCCTATCCATATGAGGCAACAGAAGAATCCTGTAAGTACA
ATCCCAAGTATTCTGTTGCTAATGACACCGGCTTTGTGGACATCCCTAAGCAGGAGAAGGCCCTGATGAAGGCAGTTGCA
ACTGTGGGGCCCATTTCTGTTGCTATTGATGCAGGTCATGAGTCCTTCCTGTTCTATAAAGAAGGCATTTATTTTGAGCC
AGACTGTAGCAGTGAAGACATGGATCATGGTGTGCTGGTGGTTGGCTACGGATTTGAAAGCACAGAATCAGATAACAATA
AATATTGGCTGGTGAAGAACAGCTGGGGTGAAGAATGGGGCATGGGTGGCTACGTAAAGATGGCCAAAGACCGGAGAAAC
CATTGTGGAATTGCCTCAGCAGCCAGCTACCCCACTGTG
Now we add the linker (first and last row):
GTACAAAAAAGCAGGCTCCACCATG
AATCCTACACTCATCCTTGCTGCCTTTTGCCTGGGAATTGCCTCAGCTACTCTAACATTTGATCACAGTTTAGAGGC
ACAGTGGACCAAGTGGAAGGCGATGCACAACAGATTATACGGCATGAATGAAGAAGGATGGAGGAGAGCAGTGTGGGAGA
AGAACGTGAAGATGATTGAACTGCACAATCAGGAATACAGGGAAGGGAAACACAGCTTCACAATGGCCATGAACGCCTTT
GGAGACATGACCAGTGAAGAATTCAGGCAGGTGATGAATGGCTTTCAAAACCGTAAGCCCAGGAAGGGGAAAGTGTTCCA
GGAACCTCTGTTTTATGAGGCCCCCAGATCTGTGGATTGGAGAGAGAAAGGCTACGTGACTCCTGTGAAGAATCAGGGTC
AGTGTGGTTCTTGTTGGGCTTTTAGTGCTACTGGTGCTCTTGAAGGACAGATGTTCCGGAAAACTGGGAGGCTTATCTCA
CTGAGTGAGCAGAATCTGGTAGACTGCTCTGGGCCTCAAGGCAATGAAGGCTGCAATGGTGGCCTAATGGATTATGCTTT
CCAGTATGTTCAGGATAATGGAGGCCTGGACTCTGAGGAATCCTATCCATATGAGGCAACAGAAGAATCCTGTAAGTACA
ATCCCAAGTATTCTGTTGCTAATGACACCGGCTTTGTGGACATCCCTAAGCAGGAGAAGGCCCTGATGAAGGCAGTTGCA
ACTGTGGGGCCCATTTCTGTTGCTATTGATGCAGGTCATGAGTCCTTCCTGTTCTATAAAGAAGGCATTTATTTTGAGCC
AGACTGTAGCAGTGAAGACATGGATCATGGTGTGCTGGTGGTTGGCTACGGATTTGAAAGCACAGAATCAGATAACAATA
AATATTGGCTGGTGAAGAACAGCTGGGGTGAAGAATGGGGCATGGGTGGCTACGTAAAGATGGCCAAAGACCGGAGAAAC
CATTGTGGAATTGCCTCAGCAGCCAGCTACCCCACTGTG
TAGGACCCAGCTTTCTTGTAC
Now we have the first problem: There is a stop codon in the 3'-linker (immediately in the beginning of the last row) even though the clone is according to the information that we received "without stop codon".
We have sequenced the clone and determined that the only difference between the with and without stop codon clones is a mutation, that converts the TAG stop codon into a TTG (leucin) codon.
So we change one A nucleotide in the sequence above into a T nucleotide:
GTACAAAAAAGCAGGCTCCACCATG
AATCCTACACTCATCCTTGCTGCCTTTTGCCTGGGAATTGCCTCAGCTACTCTAACATTTGATCACAGTTTAGAGGC
ACAGTGGACCAAGTGGAAGGCGATGCACAACAGATTATACGGCATGAATGAAGAAGGATGGAGGAGAGCAGTGTGGGAGA
AGAACGTGAAGATGATTGAACTGCACAATCAGGAATACAGGGAAGGGAAACACAGCTTCACAATGGCCATGAACGCCTTT
GGAGACATGACCAGTGAAGAATTCAGGCAGGTGATGAATGGCTTTCAAAACCGTAAGCCCAGGAAGGGGAAAGTGTTCCA
GGAACCTCTGTTTTATGAGGCCCCCAGATCTGTGGATTGGAGAGAGAAAGGCTACGTGACTCCTGTGAAGAATCAGGGTC
AGTGTGGTTCTTGTTGGGCTTTTAGTGCTACTGGTGCTCTTGAAGGACAGATGTTCCGGAAAACTGGGAGGCTTATCTCA
CTGAGTGAGCAGAATCTGGTAGACTGCTCTGGGCCTCAAGGCAATGAAGGCTGCAATGGTGGCCTAATGGATTATGCTTT
CCAGTATGTTCAGGATAATGGAGGCCTGGACTCTGAGGAATCCTATCCATATGAGGCAACAGAAGAATCCTGTAAGTACA
ATCCCAAGTATTCTGTTGCTAATGACACCGGCTTTGTGGACATCCCTAAGCAGGAGAAGGCCCTGATGAAGGCAGTTGCA
ACTGTGGGGCCCATTTCTGTTGCTATTGATGCAGGTCATGAGTCCTTCCTGTTCTATAAAGAAGGCATTTATTTTGAGCC
AGACTGTAGCAGTGAAGACATGGATCATGGTGTGCTGGTGGTTGGCTACGGATTTGAAAGCACAGAATCAGATAACAATA
AATATTGGCTGGTGAAGAACAGCTGGGGTGAAGAATGGGGCATGGGTGGCTACGTAAAGATGGCCAAAGACCGGAGAAAC
CATTGTGGAATTGCCTCAGCAGCCAGCTACCCCACTGTG
TTGGACCCAGCTTTCTTGTAC
The last operation is to insert this sequence into the empty pENTR221 sequence that we have opened in SnapGene. Practically you select the 32 nucleotides from 652 to 687 and replace them with the sequence above. Voila! Unfortunately, the fact that the linker are not always correctly indicated gives me a bad feeling. However, according to our own experience the library replicas of the Orfeome contain sufficient errors that it is anyway advisable to sequence the complete insert using T7 or M13 rev primers from the 3'-end and M13 fwd primer from the 5'-end. This way, you will figure out any linker mistakes that have been done in the annotation of the clones.