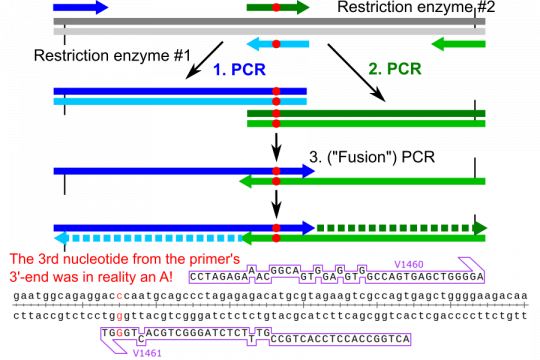
An impossible overlap-extension PCR
A PhD student of mine asked me about plasmid maps for several DNA constructs that I had created some 20 years ago. Since SnapGene did not exist at the time, I had used GeneConstructionKit2. I performed many clonings at the time, but I did not continue generating maps for all of them. The GCK2 format does not allow for easy annotation. You needed a separate program for comprehensively annotating the plasmids, and this data was saved in a separate file, the "illustration" file: what could possibly go wrong? Yesterday, I ended up digging out my old lab notebooks and retracing about 10 clonings in SnapGene. However, I was unable to simulate one of the assemblies because SnapGene was too conservative in disallowing "bad" PCR primers to function.
I had performed overlap-extension PCR to introduce a mutation into the mouse VEGF-D cDNA. The homologous mutation had been introduced into human VEGF-D before, and I therefore had the primers for the human sequences. Mouse and human VEGF-D are very similar. The primers designed to amplify the human PCR were not perfect when using mouse cDNA as a template, but none of the differences would result in amino acid changes. So I attempted the PCR with a primer that had a mismatch in the third nucleotide from the 3'-end. The PCR was successful, but even when I lowered the hybridisation parameters to the least stringent settings, SnapGene would not anneal this primer to my template. To simulate cloning in SnapGene and generate a map, I needed to introduce a mutation into my primer and then reverse the mutation after the overlap extension PCR. I guess I need to file a bug report (or would this be a feature request?). It seems appropriate that the program should be able to allow annealing of primers that do anneal in reality...