2-week Lab Course
I had no idea how much work it is to organize a practical lab course. Had I known, Pirjo would have had a much harder time to convince me to give this course for the DPBM graduate school. Kari had warned me...
The accompanying lecture course had been running from September to November. The practical course had been offered with 16 free slots, but that was totally unrealistic given that we were confined to my 23.7 square meters of lab space. Teaching lab space is available, but without equipment and all the other infrastructure that is needed for such an undertaking. 8 people registered to the practical course and - luckily - half of those pulled out in the last moment with insufficient possibility to commit to the heavy workload that the course required. Thus we ended up with four students and three projects. Under no circumstances would we have managed with more.
The idea was to offer each participant the possibility to realize his own DNA cloning and protein expression project. Something that would be relevant for his own PhD studies. For that matter, I had meetings with the three groups one month in advance to plan the cloning and to order the necessary materials. We were working in parallel on the following three projects:
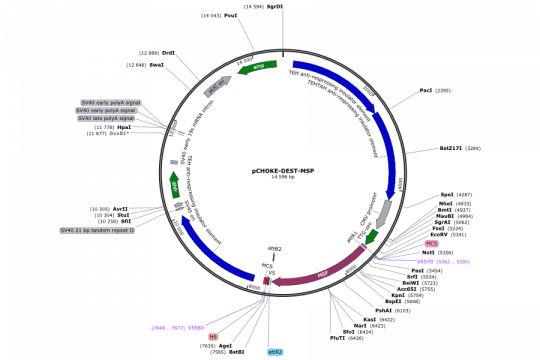
- Expression of MSP1 in CHO cells (both transient and stable)
- Expression of human AR in 293T cells (transient)
- Generation of a plasmid, that can be cleaved by a restriction enzyme into a perfectly evenly spaced DNA ladder
When I went to City Lab with my shopping list, Thermo Fisher had a sales exhibition (-33% for all enzymes). I did place the order with Fisher Scientific, clearly telling the sales rep that I would need the products one week before the start of the course and the sales rep confirmed that it would be absolutely no problem to get all items delivered in time. In reality the delivery was split up into two parts: the discounted items versus the non-discounted items. While I did place orders on both parts, only the discounted items managed to make it into Thermo Fisher’s internal ordering system due to a mistake at Thermo Fisher’s end. When the delivery came, all items were placed into the freezer/fridge, but nobody thought to check whether all the items had been delivered. When I checked on the weekend before the course, whether everything had arrived that we had ordered, I realised that stuff was missing… And I started running around trying to make sure we were able to borrow all the material that had not arrived yet. It took Thermo Fisher until December 10th to get all the items delivered. By then, we had done most of what we had planned to do.
Another bummer was, that Dharmacon/GE Healthcare was unable to deliver the human androgen receptor cDNA (BC132975, Clone ID: 40146997) from the Mammalian Gene Collection. GE Healthcare says on its pages “Ships to North America in 2 day(s) (Add one week outside of NA)”. However, even after twice that time had passed, the delivery had not happened and a few days before the course start, I was frantically trying to make up a replacement project. I managed to get a rat androgen receptor cDNA from Päivi Pihlajamaa and on the last Friday before the course Tapio still sequenced the construct junctions. The construct appeared clonable; I did a DNA prep over the weekend and redesigned the cloning plan. GE Healthcare was very apologetic for their inability to deliver and promised to deliver the human androgen receptor free of charge (but has not done so up to this moment)…
The course schedule was very tight and did not allow for any mistakes. Several times I thought that a project would end prematurely. I still find it remarkable that projects 1 and 2 worked as planned throughout the end. The closest shave was already in cloning step 1 of project 1: The cloning was non-directional and we picked many clones to be sure to find a good orientation. 9 clones had an insert, 8 in the wrong orientation and one (clone 4) in the right orientation. In the same cloning step, we inserted a chloramphenicol resistance, but the students happily plated on ampicillin plates, while they could have reduced the background noise to zero by using ampicillin/chloramphenicol plates. From the 40 colonies that they had to pick for the minipreps, only about 10 did grow when we used both antibiotics for the culture.
The transfection and detection went smoothly without any problems. For the final imaging, we used Licor's Odyssey Fc, which our Research Program had acquired a few weeks earlier and I was surprised about its high sensitivity (I took the ECL membrane and exposed it to X-ray film to get a feeling of how they compare).
We had no choice to halt project 3 after the first two cloning steps (out of 3). When we checked the sequences of the construct pOTB7-BglI2x-insert, we found a mutation in one of the BglI overlaps (this mutation does not affect cleavage but does affect compatibility for ligation). We sequenced another two constructs, but all of them showed the same mutation and we realised that the originating construct (pOTB7-PC5) contained already this mutation. Because we rely in the last cloning step on the specific overhang generated by this BglI site, we could not continue short of fixing this mutation. We ordered the mutagenesis oligonucleotides immediately, but for this course this project was over. Because Girma is still invested in finishing this project, we will fix the problem and do the missing last cloning step. It would be nice to have your own evenly spaced DNA marker made by restriction digest of a DNA construct. That’s what everybody used 25 years ago: Lambda HindIII or something similar. Only the fragment spacing was never really good. In our incarnation of the evenly spaced DNA ladder, we can clone a HindIII/SpeI fragment into our base vector (pOTB7-BglI2x) and in a second concatenating ligation step the length of the HindIII/SpeI fragment determines the size-spacing of the resulting ladder. During this course we used a 350bp fragment, which would have given us a DNA ladder like this: 350, 700, 1050, 1400, 1750, 2100, etc.).
Once we finish this construct, all students will get the plasmid and an aliquot of the ready ladder as a souvenir. Because the costs to make this ladder are about 40 times less than the buying a commercial DNA ladder, we could even start to give it away for free. Some things in the preparation of this course could have been easier: Just on the last course day, I received from Gretchen Repasky the "DPBM guide for course organizers". Why did I never got to know that somethings like that exists????? Johanna Timmerbacka, the previous graduate school coordinator, must have wondered the same: I asked her via e-mail all the questions that this guide answers...
Attached to this story are the cloning plans and the schedule for the course. The collection of protocols and a short description of the results will be added asap after the course. There are also two references (one from the US patent office and one from the Journal of Nucleic Acids) about two alternative ways how to create DNA ladders. Bottom line: Next year we won't do anything wet. We'll sit in front of computer screens. Maybe. If we find a teaching class in Biomedicum with a beamer that has a resolution higher than 1024x768.