SnapGene and partial restriction digests revisited
Snapgene is a software for the wet lab molecular biologist, who does lots of cloning work (construct design and annotation). Since I last wrote about the SnapGene software (https://www.snapgene.com/), many good things have happened:
- The Snapgen Server: http://www.snapgene.com/products/snapgene_server/. If you want to publish your constructs online, this would be a nice way to go. I tried it out and it's fairly easy to deploy.
- Support for protein sequences: This is still experimental and many options are grayed out (at least on my version). SnapGene focuses on implementing features fast and the documentation often lags behind. For me, this feature is not essential since I do most of my protein sequence work with BioPython or EMBOSS. However, I can see that this would be useful for wet lab scientists who quickly want to get some work done without spending hours on writing a Biopython script or learning the syntax of EMBOSS commands.
- Support for collections: A collection is simply a folder with SnapGene files. The program is able to read all files within that folder and you can perform actions on the collection (e.g. search for a specific sequence).
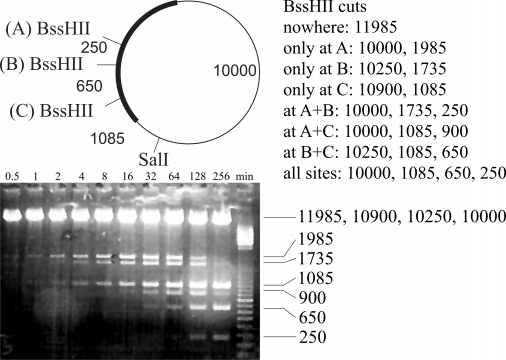
However, Snapgene has still not implemented a feature that we would like to have: partial restriction enzyme digests. This comes in handy when you do an analytical digest and you did not add enough enzyme. Or the enzyme was already mostly dead. Then you get typically a complex pattern, from which you very often can deduce (with some effort), whether your construct is OK or not. However, this requires manually calculating all possible fragment lengths, which can be quite some work if you have used e.g. two enzymes, which both cut your DNA at multiple sites. This is exactly the type of work that computers are ideal for! Sometimes, you also want to do a partial digest on purpose. For example, if you want to isolate an insert that can be excised only with a restriction enzyme that cuts also within your insert as shown in the image: Despite two additional BssHII cuts within the sequence, it is possible to isolate the 1985-bp-long BssHII-SalI fragment.
Many companies and labs have started to use SnapGene (many of them based on our recommendations). Among them is Addgene, a very useful nonprofit distribution channel for DNA constructs (https://www.addgene.org/news/addgene-upgrades-plasmid-maps-snapgene-serv...). If you are considering to purchase a license, you get a 10% discount when using my email address (michael@jeltsch.org) as a discount code.
UPDATE (17.07.2020): SnapGene is changing their licensing scheme (no permanent licenses anymore, only yearly subscription fees). Also referral discount codes have been discontinued (hence the strikethrough text above.